Viral Classification
The usual means to classify and identify living things is to use the binomial system of naming devised by Carolus Linnaeus.
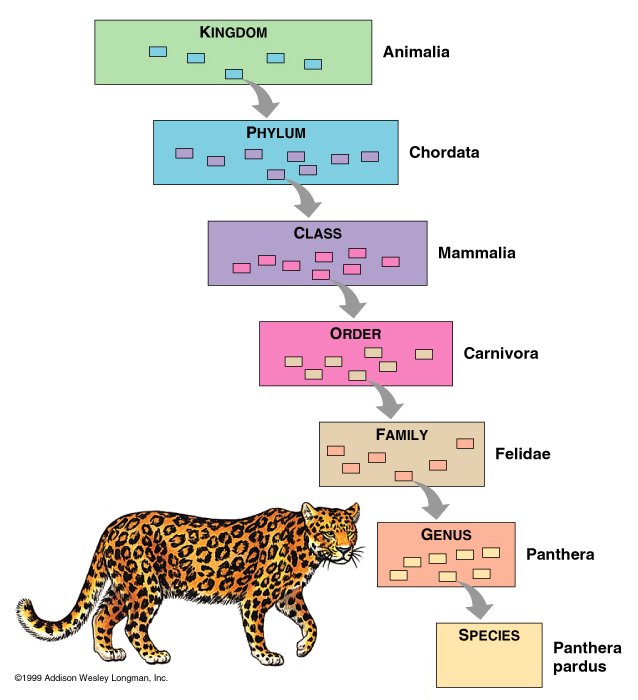
The hierarchy of classification:
http://www.mun.ca/biology/scarr/138599_KPCOFGS.jpg
One good mnemonic is: Keep Plates Clean Or Family Gets Sick!
However, because viruses are so different from cellular organisms, it is difficult to classify them according to the typical taxonomic categories (i.e. Kingdom, phylum). Instead, viruses may be classified with the ICTV classification, Lwoff’s scheme of classification, or the Baltimore’s system of classification.
ICTV Classification
· Single, universal taxonomic scheme for viruses.
· Devised by the International Committee on Taxonomy of viruses (ICTV).
· Family is the highest taxonomic category used.
· Criteria includes:
o Nature of genome & sequence relatedness
o Virus structure
o Natural host range
o Cell and tissue tropism
o Pathogenicity and cytopathology
· Common name, rather than binomial term used to designate a viral species.
· Problems such as distinguishing between viral species and viral strains have not been resolved completely.
Example:
Nidovirales (Order)àCoronaviridae (Family)àCoronavirus (Genus)àInfectious Bronchitis Virus (type species)
Note: A type species is a species whose name is linked to the use of a particular genus name. The genus is so typified will always contain the type species.
Lwoff’s Scheme of Classification
· Devised in 1962 by Lwoff, RW Horne and P Tournier
· Based on the physical properties of the virus:
o Nucleic acid
o Symmetry of capsid
o Presence/absence of envelope
o Dimensions of virion and capsid
Baltimore’s System of Classification
· Devised by American biologist David Baltimore.
· Encompasses all viruses
· Based on the viral genome and its relationship to mRNA, modes of replication and gene expression.
· Can make inferences and predictions about the fundamental nature of all viruses within each defined group.
· Baltimore’s 1971 Paper titled “Expression of Animal Virus Genomes” : http://mmbr.asm.org/cgi/pmidlookup?view=long&pmid=4329869
There are 7 classes in the Baltimore Classification.
Class I (dsDNA)
Adenoviruses, Herpesviruses, Poxviruses, Papillomaviruses, Mimiviruses.
Class II (ssDNA)
Parvoviruses, Inoviruses, Microviruses, Anelloviruses, Nanoviruses.
Class III (dsRNA)
Reoviruses, Cystoviruses, Birnaviruses, Totiviruses, Partitiviruses.
Class IV ((+)-sense ssRNA)
Picornaviruses, Togaviruses, Flaviviruses, Astroviruses, Barnaviruses.
Class V ((-)-sense ssRNA)
Orthomyxoviruses, Paramyxoviruses, Rhabdoviruses, Filoviruses, Bunyaviruses.
Class VI (ssRNA reverse transcribing)
Retroviruses, Metaviruses, Pseudoviruses
Class VII (dsDNA reverse transcribing)
Hepadnaviruses, Caulimoviruses