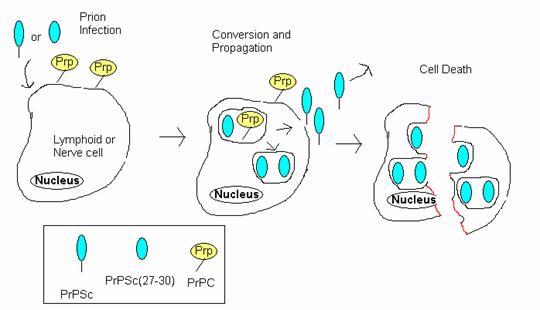
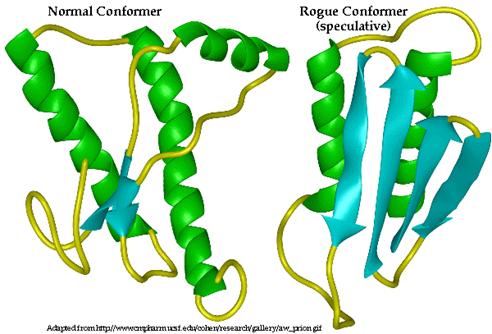
Isolation and Cultivation of Viruses
Animals and eggs can be used for virus cultivation. However, this way of cultivating viruses has been mainly replaced by cell culture due to the inconvenience and safety involved in handling animals. Animals and eggs still have their advantages for use as a culture “media”—this method is still used to cultivate viruses that have no known host in vitro (e.g. influenza virus in chick embryo)), and to study viral pathogenesis in a whole host (e.g. polo studies in chimpanzees). Plants may also be used to cultivate certain plant viruses. The tobacco mosaic virus may be used to determine virus numbers based on the number of virus plaques on its leaves.
Cell tissue culture comprises of cells being grown in vitro on continuous cell lines. Continuous cells lines are the preferred choice of viral culture because they can be sub-cultured indefinitely, theoretically. They are derived from polyploid or multiploid cancerous cells. Viruses that do not grow in vitro have to be grown in animals, plants or eggs.
Detection, identification and diagnosis (elaborated later)
1. Tissue culture methods
a. Cytopathic effect
b. Plaque assay
2. Physical methods
a. X-ray crystallography
b. Electron microscopy
c. Ultracentrifugation
3. Serological methods
a. Haemagglutination (HA)
b. Haemagglutination Inhibition (HI)
c. Virus neutralisation
d. Complement Fixation
4. Immunological methods
a. Immunofluorescence
b. Immunogold EM
c. Immunoprecipitation
d. Immunoblot
e. Enzyme Linked Immunosorbent Assay (ELISA)
5. Molecular biology methods to analyse viral proteins
a. SDS PAGE
b. Western blot
c. Protein sequencing
d. X-ray crystallography
6. Molecular biology methods to analyse viral genome
a. Restriction analysis
b. DNA sequencing
c. Southern blot
d. Northern blot
e. PCR/RT-PCR
Cytopathic Effect:
Cytopathic effect (CPE) is an alteration in cell morphology resulting from viral infection of a cell culture monolayer. CPE may be used as a presumptive identification of a virus. If the virus does not produce a CPE, its presence can be detected by several other techniques such as haemadsorption.

http://www.microbelibrary.org/microbelibrary/files/ccImages/Articleimages/cummings/baculovirus.JPG
Plaque Assay:
Plaque Assays are used for the quantitative measure of infectious centres by counting the number of infectious virus particles in a given sample in Plaque forming units.
The plaque assay is based simply on the ability of a single infectious virus particle to give rise to a macroscopic area of cytopathology (i.e. plaque, focus of infection) on an otherwise normal monolayer of cultured cells.
Specifically, if a single cell in a monolayer is infected with a single virus particle, new virus resulting from the initial infection can infect surrounding cells which, in turn, produce virus that infects additional surrounding cells. Multiple rounds of infection give rise to an area of infection called a plaque.
*The semisolid medium prevents the formation of secondary plaques through diffusion of virus from the original site of the infection to new sites, ensuring that each plaque that develops in the assay originated from a single infectious particle in the starting inoculum.

http://www.v-bio.com/pictures/p06_small.png
Advantages
1. Simple, inexpensive, precisely quantitative method.
2. Higher accuracy at lower concentration—useful for samples with very low virus counts (in food and water samples).
Disadvantages/Limitations
1. Counts only viable virus (virus capable of multiplying)
2. Only works for viruses that infect monolayer cells and viruses that cause cell lysis (or other cytopathic changes that can be observed)
3. Must know culture conditions for the virus studied.
4. Requires time for incubation, time consuming.
Calculation
Original Virus Concentration (in PFU/ml) = Final Virus Concentration * Dilution Factor * (1/volume pipetted in well (in ml))
X-ray crystallography:
Viruses are crystallized and by using X-ray diffraction techniques, the structure of the virus can be elucidated from the diffraction pattern produced.
Electron microscopy:
This includes Transmission EM, Scanning EM and STEM. Electron microscopy involves scattering electrons on a specimen to “illuminate” it and magnify it for viewing. The resolution of an electron microscope is much greater than a light microscope as the wavelength of an electron is much smaller than that of a light proton.

http://www.biologie.uni-hamburg.de/b-online/ge03/14.gif
Ultracentrifuation:
Ultracentrifugation has assumed growing importance in virus diagnosis as a technique by which to concentrate and purify viruses for immediacy diagnosis on the basis of electron microscopy as well as for purely virological and serological tests.
Ultracentrifugation has proved to be helpful for sizeable improvement of sensitivity for detection, which, in turn, has been conducive to time saving. The preparational ultracentrifuge enables also direct diagnosis by determination of isodensities and sedimentation coefficients of viruses and their components.
Haemagglutination:
Agglutination of red blood cells caused by certain antibodies, virus particles or high molecular weight polysaccharides. This can be used to test for influenza and other viruses with two spike proteins neuraminidase and haemagglutinin, since these two proteins bind specifically to red blood cells.
Haemagglutination Inhibition:
This is used to quantify the virus by haemagglutination. If the virus and antibody are homologous, the antibody bound to the surface of the virus blocks its entry into the cell. This neutralizes infectivity, because it prevents viral replication and subsequent CPE formation or animal infection. Agglutination is also i
nhibited.

http://www.umanitoba.ca/science/microbiology/staff/cameron/graphics/401lab3passivehemagglutination%20group506.jpg
Virus neutralization:
If the virus and antibody are homologous, the antibody bound to the surface of the virus blocks its entry into the cell. This neutralizes viral infectivity, because it prevents viral replication and subsequent CPE formation or animal infection. It is one of the methods employed for the detection of virus-specific neutralizing antibodies.
Complement fixation:
Complement fixation is used to detect the presence of either specific antibody or specific antigen in a patient's serum. If the antigen (the unknown virus in the culture fluid) and the known antibody are homologous, complement will be fixed (bound) to the antigen-antibody complex. This makes it unavailable to lyse the “indicator” system, which is composed of sensitized red blood cells.
Immunofluorescence:
Microscopic method of determining the presence or location of an antigen (or antibody) by demonstrating fluorescence when the preparation is exposed to a fluorescein-tagged antibody (or antigen) using ultraviolet radiation.

http://www.microbelibrary.org/microbelibrary/files/ccImages/Articleimages/cummings/1245-MERGED%2020x%20wide%20field.JPG
Above: Immunofluorescence for Herpes Simplex Virus Antibody
Immunogold EM:
Locate specific proteins or antigens by attaching nano-gold particles to antibodies.
Immunoprecipitation:
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins. Immunoprecipitation requires that the antibody be coupled to a solid substrate at some point in the procedure.
Immunoblot:
The immunoblot (also called Western blot) is an analytical technique used to detect specific proteins in a given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native or denatured proteins by the length of the polypeptide (denaturing conditions) or by the 3-D structure of the protein (native/ non-denaturing conditions). The proteins are then transferred to a membrane (typically nitrocellulose or PVDF), where they are probed using antibodies specific to the target protein.
ELISA (Enzyme-linked Immunosorbent Assay):
The basic principle of an ELISA is to use an enzyme to detect the binding of antigen (Ag) and antibodies (Ab). Antibodies are bonded to enzymes; the enzymes remain able to catalyze a reaction that converts a colourless substrate to a coloured product, indicating the presence of Ag:Ab binding. Depending on the test, an ELISA can be used to detect the presence of either Abs or Ags.

https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEinBG2coFfuUX6aMFfIan1riPrk5KyKN8LrZiUjosvF0yIXlwJpVz4X9XDVZR9fLa9HUKOAl50exCkNaYjJvCW_udpAJqsFFTYu9600FRnzfsu6Vh1U-o3tTcu3fwdLKE6szx7qAzra5VAC/s1600-h/kakia.jpg
SDS PAGE:
SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis, is a technique widely used in biochemistry, forensics, genetics and molecular biology to separate proteins according to their electrophoretic mobility.
Polymerase Chain reaction (PCR):
A method for expanding small discrete sections of DNA by binding DNA primers to sections at the ends of the DNA to be expanded and using cycles of heat (to create single-stranded DNA) and cooler temperatures (to allow a DNA polymerase enzyme to create new sections of DNA between the primer ends).

http://www.copernicusproject.ucr.edu/ssi/HighSchoolBioResources/Genetic%20Engin%20Hum%20Genome/pcr.jpg
Southern Blot:
Identification of specific genetic sequences by separating DNA fragments by gel electrophoresis and transferring them to membrane filters in situ. Labelled complementary DNA applied to the filter binds to homologous fragments, which can then be identified by detecting the presence of the labelled DNA in association with bands of certain molecular size. This test was named after its discoverer, E.M. Southern.

http://www.biogate.it/microarrays/images/Southern_blot.gif